A word from the Director
The exposome is composed of four entities: chemical, physical, biological, and social. The concept was introduced in 2005 by Chris Wild, encompassing all life-course environmental exposures, complementing the genome. Several contributions expanded on this idea, incorporating lifestyle factors, internal chemical exposures, and biological responses. Computational approaches highlighted the importance of modeling and biological connectivity. Currently, the exposome is studied on a large scale with the aim of better understanding the links between exposures and their health effects, combining different types of exposures and monitoring them more extensively.
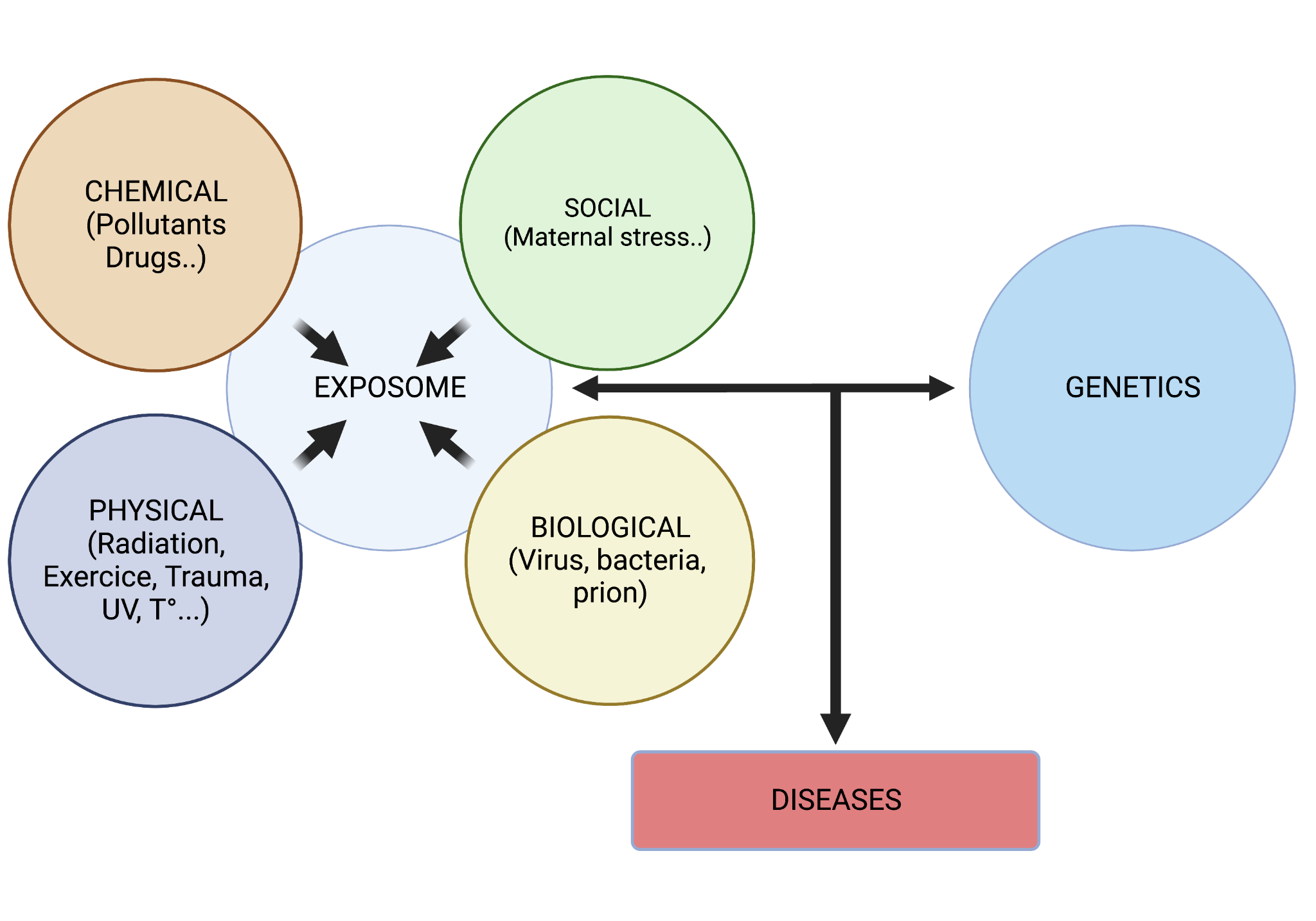
The components of the exposome and their interactions with genetic factors, leading to the development of pathologies.
Our Inserm/UPCité research unit “Health and functional exposomics” (HealthFex) is entirely in line with this strategy by focusing on the consequences of the exposome and the mechanisms of exposome compounds on health. We favor a universal definition of the term “functional exposomics” to embody a practical approach to advance understanding of all external factors contributing to the phenome (i.e., the totality of traits/characteristics displayed by a biological entity). Yet we will focus on certain outcomes to ensure efficiency.
Functional exposomics is the systematic and comprehensive study of environmental exposure-phenotype interaction over a defined time-period. The projects carried out by the unit will aim at understanding the complex interrelationships of exposure and endogenous processes. In analogy to functional genomics that focused on the biological functions linked to genes, functional exposomics focuses effort on those exposures that have a demonstrable impact on phenotype.
Our unit will be composed of six teams working on different components of the exposome:
- Team 1: Metatox – Metabolism, cell signaling, toxicology and biomarkers (Xavier Coumoul / Etienne Blanc / Emmanuelle Bouzigon)
- Team 2: Systox – System biology and toxicology (Karine Audouze)
- Team 3: Myelination and system nervous pathologies (Charbel Massaad)
- Team 4: Degeneration and plasticity of the locomotor system (Frédéric Charbonnier)
- Team 5: Addictions and comorbidities: biomarkers, mechanisms and pharmacotherapies (Florence Noble)
- Team 6: Cell death in host-pathogen interactions (Jérôme Estaquier)
Positioning of the project in the national or international scientific field: the position and visibility of the unit at the national level, European and international levels will still be central in terms of research on functional exposomics. We believe that the coherent global project will support our leadership in the field. Although we are still working on delineating the roles of various signaling pathways in multiple pathologies, the vision of the new project will now address the interplays of the different components of the exposome and thus support the emergence of new projects within the unit with external partners, incorporating multiple exposures to better characterize how they influence human health. This evolution has already started in several teams which used to study simple stress (e.g., single pollutants) and now focus on complex mixtures or on interaction of chemicals with psycho-social stressors. The orientations of the research unit represent:
- academic research (production of new knowledge).
- transfer & outreach activities (valorization of research findings, expertise, transfer, dissemination).
- support activities (at the service of the academic or scientific communities).
Our projects envision the use of resources currently available (e.g., within the unit, type 2 culture facility, radioactivity and microbiology confinement laboratories (specific rooms, technical facilities of the UFR Sciences Fondamentales and Biomedicales, Faculté des Sciences, and the potential future Federation with the CNRS BFA unit of Université Paris Cité).
We are also part of a national exposome facility (France exposome) and European ESFRI on the exposome (EIRENE) and on computational sciences (ELIXIR)
